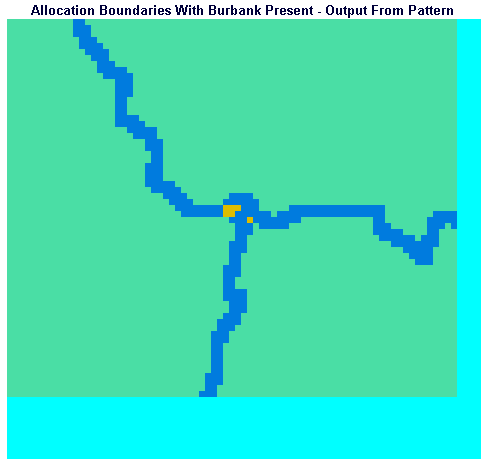
In order to determine the number of people within a specific shared buffer zone, e.g., the three-minute buffer between Burbank Hospital and Nashoba Hospital catchments, it is necessary to differentiate the portions of the buffer.
This will be demonstrated using IDRISI for the scenario in which Burbank Hospital exists. It is left to the reader to work through the process for the Burbank Absent scenario, if desired.
Display the image ALLOWBND using the QUAL256 palette. This image shows the hospital catchment boundaries as pixels with a value of two and threes on a background of ones. Recall that this image was created by the PATTERN module which placed in each pixel the number of different pixel values in the eight neighboring pixels. Wherever two catchment areas, whose pixels had different values, abutted, PATTERN would place a two in the cell. Where three catchment areas abutted, PATTERN placed a three. Notice that there are four junctions in the image, corresponding to places where three catchment areas meet.
We are going to use the GROUP module to differentiate the segments of the boundary lines between catchments. GROUP will not differentiate between segments if the pixels making up the segments share edges contiguously.
If the Idrisi Toolbar is not visible, turn it on by checking the Toolbar option in the Environment menu. Click on the Window icon, which looks like a pair of nested rectangles just to the right of the Cursor Inquiry (question mark) icon. Using the Window tool, examine each of the four junctions.
We are going to use the GROUP module to differentiate the segments of the boundary lines between catchments. GROUP will not differentiate between segments if the pixels making up the segments share edges contiguously.
If you window in closely on the northernmost (top) junction in the ALLOWBND image, you will notice that the branch which leaves the junction toward the west (left) is isolated by the pixels with a value of three at the junction. However, the branches which come in from the east (right) and south (bottom) are not separated.

The next junction as you move southward (downward) in the image has effective separation between the branches. The third junction is problematic as the first was. The branch leaving the junction toward the south is separated, but the branches leaving toward the east and northwest are not separated.
We will manually alter the pixel values at the two problem junctions to enable GROUP to give us the result we require, namely separating the segments of the catchment boundaries.
From the File menu run the FILE MAINTENANCE module. Choose to copy the image file ALLOWBND to create the new file BOUNDS.
Run UPDATE from the Data Entry menu. Specify an Input Image of BOUNDS. Indicate that you wish to place a value of one in rows 155 to 159 and columns 538 to 542. Click the right arrow to specify a second range, indicating a new value of one from rows 496 to 499 and columns 428 to 430.
Display BOUNDS using the QUAL256 palette and window in on the top junction to see that the boundary branches are now separated. Do the same for the third junction.
Now we are ready to run GROUP from the Context Operators section of the Analysis menu. Be sure to UNCHECK the Include Diagonals box. Specify an Input Image of BOUNDS and an Output image of BOUNDGRP.
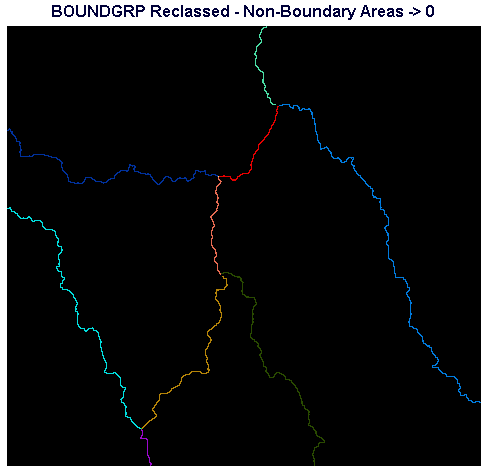
Inspect BOUNDGRP using the QUAL256 palette. If you window in on the segments of boundaries you should observe that they are now separated into discrete branches. Use Cursor Inquiry to note the pixel values in the large catchment areas. We need to run RECLASS to set these values to zero. Only the boundaries themselves should have non-zero values.
Assuming the values you observe for the non-zero catchment areas which should be changed to zero are 3,6, and 10, run RECLASS from the Database Query section of the Analysis menu and specify that you wish to use BOUNDGRP as the input image and BNDSDIFF as the output image. Specify that new values of zero should replace all values from 3 to just less than 4, 6 to just less than 7, and 10 to just less than 11.
Display BNDSDIFF using the QUAL256 palette. If all went well with the preceding steps, only the boundary lines are visible, and each is in a different color.
We need to create a new travel time map using the new, segmented catchment boundaries as the origin of travel. Run COST from the Distance Operators section of the Analysis menu. Use BNDSDIFF as the Feature Definition Image and PRCLF14 as the Friction Surface Image to create BOUNDSTT.
Run ALLOCATE from the Distance Operators section of the Analysis menu. Specify BOUNDSTT as the Distance Image, BNDSDIFF as the Target Image, and BOUNDALO as the Output Image.
BOUNDALO represents the allocation of the entire study area (including areas beyond three minutes from the boundaries) to catchment boundary segments. We need to limit the allocation polygons to those areas within the three-minute buffers. One easy way to do this is to do a multiply overlay of BOUNDALO with the BUFFWB buffers image we created before.
Display BUFFWB using a qualitative palette. If you use Cursor Inquiry to check pixel values, you should observe that the areas outside the buffers have a value of zero and areas in the buffers have a value of 10. We assigned the buffers a value of 10 to facilitate interpretation of the EXTRACT steps at the end of Section E, above. We can still use the BUFFWB image for the present step.
Run OVERLAY from the Database Query section of the Analysis menu. Specify that you wish to perform a multiplication (*) overlay between BOUNDALO and BUFFWB. Call the Output Image PAIRBUFF.
Display PAIRBUFF with the QUAL256 palette. Note that pixels in each buffer segment have different values. The final step is to run EXTRACT to sum the populations within each segment of buffer.
Run EXTRACT from the Database Query section of the Analysis menu, specifying PAIRBUFF as the Feature Definition Image and POPDENS as the Image TO Be Processed. Select the Sum option and Tabular Display.
EXTRACT produces this output:
Population Within Specific Buffer Segments
| Category | Total |
|---|---|
| 0 | 318970.186696 |
| 10 | 1558.884606 |
| 20 | 6747.807235 |
| 40 | 1020.167243 |
| 50 | 1723.416872 |
| 70 | 0.341945 |
| 80 | 694.943929 |
| 90 | 3454.424131 |
| 110 | 4165.366264 |
| 120 | 8369.466488 |
| 130 | 6.183905 |
| 140 | 1449.544993 |
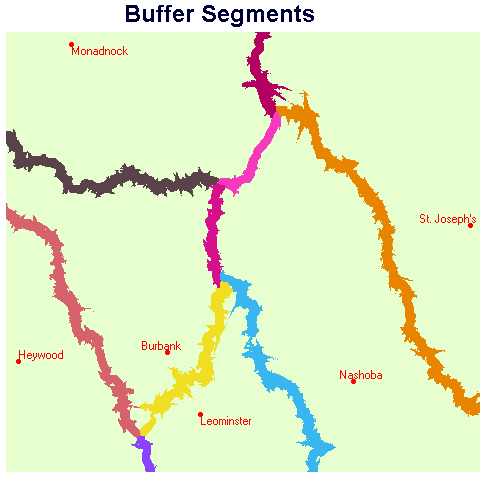
Display PAIRBUFF using the BUFFERS palette. Add the vector points layer HOSPWB using the default IDRPTS symbol file. Add the vector text layer HOSPWBT using the HOSPTEXT symbol file.
In order to determine the population in a particular buffer, one needs to determine the ID of the buffer, by using Cursor Inquiry on PAIRBUFF, and then consult this table produced by EXTRACT. For example, clicking on the buffer between Monadnock and Nashoba yields a pixel value of 40. The corresponding population which lives in this buffer area is 1,020 people.
A few final comments regarding the table produced by EXTRACT are warranted. Category 0 represents the total population outside of all buffers - EXTRACT summed the pixel values in POPDENS corresponding to background (zero pixels) in the PAIRBUFF image. Category 70 corresponds to the population corresponding to anomalous pixels in the middle of the second buffer junction from the north in PAIRBUFF. This is the result of the UPDATE step which inserted values in the BOUNDS image. Category 130 represents the population corresponding to anomalous pixels in the southernmost junction. In both cases, Categories 70 and 130, the anomalies are the result of GROUP creating groups from isolated sets of pixels. These groups persisted through the cost distance and allocation stages of the analysis.
Rejoin the main study